Articles
Chapter 11: Aids to taxonomic identification
N. Maxted
School of Biological Sciences, University of Birmingham, UK
E-mail: nigel.maxted(at)dial.pipex.com
|
2011 version |
1995 version |
||
|
Open the full chapter in PDF format by clicking on the icon above. |
|||
This chapter is a synthesis of new knowledge, procedures, best practices and references for collecting plant diversity since the publication of the 1995 volume Collecting Plant Diversity; Technical Guidelines, edited by Luigi Guarino, V. Ramanatha Rao and Robert Reid, and published by CAB International on behalf of the International Plant Genetic Resources Institute (IPGRI) (now Bioversity International), the Food and Agriculture Organization of the United Nations (FAO), the World Conservation Union (IUCN) and the United Nations Environment Programme (UNEP). The original text for Chapter 11: Aids to Taxanomic Identification, authored by N. Maxted, has been made available online courtesy of CABI. The 2011 update of the Technical Guidelines, edited by L. Guarino, V. Ramanatha Rao and E. Goldberg, has been made available courtesy of Bioversity International.
Please send any comments on this chapter using the Comments feature at the bottom of this page. If you wish to contribute new content or references on the subject please do so here.
Back to list of chapters on collecting
Internet resources for this chapter
|
Practicing characterization using a color chart at a training course (photo: Bioversity) |
Abstract
To effectively conserve and utilize species, the conservationist must be able to distinguish and identify them correctly. The process of specimen identification or determination involves two steps: the decision as to which taxon (e.g., genus, species or subspecies) the specimen represents and what the “accepted” name for it is, if more than one name has been used for that taxon. The first step is achieved through some form of key or identification aid, while the second involves finding the accepted name for the taxon. Taxa are still primarily identified using single-access dichotomous keys, although multi-access keys and illustrations or photographs can be used.
In this updated chapter on taxonomic identification, progress has been reviewed in six key areas: (1) botanical glossaries, (2) combining text and illustrations for identification, (3) interactive identification, (4) conservation field guides, (5) barcode identification and (6) accepted names. Although there have been advances in identification science in recent years, poor taxonomic training and lack of user-friendly identification aids remain an inhibiting factor in the conservation and use of plant genetic resources.
Introduction
The original chapter starts with the statement that “Increasing attention is being focused on the collection and conservation of wild species” but since that text was written, the focus of conservation of plant genetic resources has turned even more towards conserving wild species, which has meant identifying greater numbers of specimens. Crop diversity is already largely conserved, at least for the major crops, and seed is conserved ex situ in genebanks. The second report of the Food and Agriculture Organization of the United Nations (FAO) on the state of the world’s plant genetic resources for food and agriculture (SoW2) reports that 90% of genebank accessions are of crop material (FAO 2010); however, there is growing recognition that crop wild relatives (CWR) are an under-exploited resource (Maxted and Kell 2009).
Feuillet et al. (2008) question the ability of breeders to increase or simply sustain crop yield and quality in the face of growing and dynamic biotic and abiotic threats. They suggest that the increasing ease of gene discovery, the development of enabling genetic and breeding techniques and a better understanding of the previous limitations on exotic germplasm make CWR the obvious choice for meeting contemporary demands for food security. This is reflected in the clear increase in the number of CWR collections between 1996 and 2009 (FAO 2010).
Given the changing focus of germplasm collection from crop to wild species (FAO 2010), the growing need to establish genetic reserves for in situ CWR conservation, and the demise of teaching taxonomy (House of Lords 2002), the need for skills in plant identification has never been greater. It is probable that lack of identification skills remains a fundamental limitation to plant genetic resource conservation and use. How can we conserve or exploit plant diversity if we cannot recognise it in the field?
Current status
With the imperative to conserve and use wild biodiversity, it might be expected that the taxonomic community would respond with a new generation of identification tools that avoid the pitfalls of classical botanical terminology and the arcane practice of using keys. But today, field identification is still largely based on the use of a hand lens and dichotomous keys: there have been no fundamental changes since 1995 (and even in fact 1895) when the original texts were written. However, some new developments that offer opportunities and challenges are noted below.
Botanical glossaries
One of the major limitations to using traditional keys in the field is the extent of the botanical terminology used. By definition, dichotomous keys are based on variations in gross morphological characteristics, and the range of characteristics used to describe the major groups of taxa will vary substantially between groups because their basic morphology is different. The suite of characteristics used to describe cereals will be very different from those describing legumes, and they will both be very different from wild cucurbit species.
Each major group of plants has its own associated descriptive terminology, but today there are very few botanists who have sufficient skills (i.e., knowledge of the general and group-specific terminology) to identify any plant species encountered in the field. Almost in recognition of this fact, there have been several botanical glossaries published in recent years: notably, Hickey and King (2000), which is split between a written glossary and a series of annotated illustrations of plant parts; Harris and Woolf Harris (2001) and Beentje (2010), which are both a glossary with illustrations throughout. It is advisable for any plant conservationist collecting from the wild, surveying a genetic reserve, or re-identifying grown-out specimens to keep one of these glossaries close to hand.
Combining text and illustrations for identification
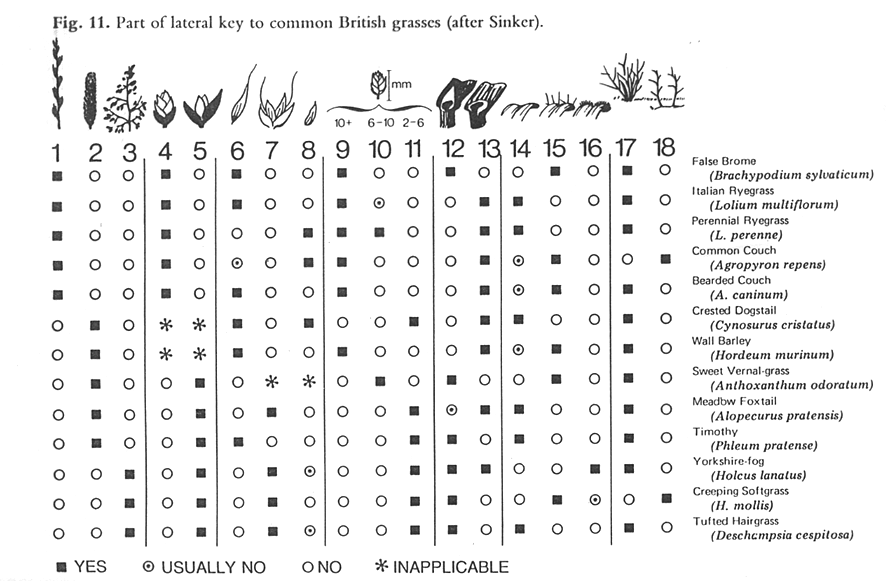
When describing the various ways to identify biodiversity, many of the taxonomic text books recommend a combination of text and images in keys to avoid reliance on botanical terminology alone. Figure 1 shows a lateral multi-access to British grasses, where the illustrations at the head of each column clearly show the characteristic described. Although this remains an efficient concept, it has been taken up by surprisingly few taxonomists. No major Floras routinely use illustrations within their keys, and the few examples where textual keys and illustrations are combined are often associated with specialist publications, such as Aids to Identification in Difficult Groups of Animals and Plants (AIDGAP) (www.field-studies-council.org/publications/aidgaptesters.asp). The AIDGAP project is producing a range of books designed to provide clearly written and illustrated guides to enable the non-specialist to identify specimens in the field and lab—techniques that could clearly be usefully applied in the context of plant genetic resource conservation.
 |
|
Source: Pankhurst (1978) Figure 11.1. Part of a lateral key to common British grasses. |
Interactive identification
The nature of the traditional dichotomous keys means that to identify a specimen, the user must follow a specific and structured sequence of questions until the identification is finally made. However, the user might not be able to proceed past the first couplet because neither of the leads provided in the first couplet fits the specimen (e.g., the first couplet might ask a question about seed characteristics but no seeds are present on the specimen) or neither of the two alternative descriptions in a couplet appears to describe a specimen accurately.
Multi-access keys were developed to overcome some of the problems associated with single-access keys. A multi-access key does not force the user to go through the character set in a specific preordained sequence; rather, it allows the user to ignore particular characters and still obtain an identification. For example, if the specimen lacks seeds, the identification can be based on vegetative and flower characteristics alone.
Computers are now commonly used for multi-access identification—a process that is referred to as “interactive identification” because the user interacts with the identification program and data set to obtain the identification. Given that identification is often an unconscious process, interactive identification keys should try to mimic the way we identify objects naturally and help reduce the user's frustration when the process becomes more explicit. Stevenson et al. (2003) suggested that these keys should (1) provide training tools and games to let people become familiar with the “cast of characters” slowly instead of being overwhelmed and confused by having to learn a lot of new things at once, (2) work to reduce the time necessary to identify a species by choosing likely possibilities from a line-up approach and (3) suggest further queries that will aid in making the final positive identification. The interactive identification program holds a matrix of taxa against characters, possibly including both text and images, and the user enters attributes (character-state values) of the specimen to be identified. The program eliminates taxa whose attributes do not match those of the specimen. This process is continued until only one taxon remains, giving a provisional identification. As with all identifications, the specimen may then be checked against a description, images or already named specimens to confirm the identification.
There are several interactive identification programs now available. For example, Linnaeus II (www.eti.uva.nl/products/linnaeus.php) is a commercial program available from the University of Amsterdam, the Netherlands. It is free, if you register as a data provider. Linnaeus II supports the creation of taxonomic databases, optimizes the construction of easy-to-use identification keys, expedites the display and comparison of distribution patterns, and promotes the use of taxonomic data for biodiversity studies. There are three modules of Linnaeus II: the “Builder” to manage your data and to create an information system, the “Runtime” engine to publish completed information systems on CD-ROM/DVD-ROM, and the “Web Publisher” to publish your completed project as a website.
A second example is Lucid (www.lucidcentral.com/), which is available from the Centre for Biological Information Technology, University of Queensland, Australia. Similar to Linnaeus II, the Lucid system consists of a number of inter-related products that assist with the creation and use of keys (in any language) for any group of organisms. Lucid software is a special type of expert system, specifically designed for identification and diagnostic purposes, which enables expert knowledge to be “cloned” and distributed to a wide audience via CD or the internet. Lucid-based keys are accessed via the Lucid Player, which provides the interface for users to load and interact with the Lucid keys, using text, images, videos and sounds to help select those taxonomic, diagnostic or other features that best describe the particular case being investigated. The key works by elimination: as the user selects character states, the program eliminates the taxa that do not possess that state. This is repeated until a single taxon remains and the identification is achieved. Once a specimen has been identified, the user is provided with a full range of multimedia fact sheets: descriptions, conservation notes, sub-keys for infra-specific taxa or links to websites for further information or recommendations. Lucid keys can be built in various languages and use terminology familiar to the user, allowing the package to be used internationally and across a wide range of capabilities.
Both the Linnaeus II and Lucid 3 identification keys are being increasingly used by a wide range of people, from high school and university students to taxonomists, quarantine identifiers, biodiversity scientists and conservation managers. As an example of an interactive key, an exemplar data set for African Vigna (Maxted et al. 2004) is included in Lucid 3 and can be downloaded from the Lucid website (http://keys.lucidcentral.org/keys/African_Vigna/default.htm).
Conservation field guides
Anyone working in field conservation is quickly forced to address the identification problem: how do I identify all the species I am confronted with in the field and wish to conserve? The majority of identification aids produced by taxonomists are, by far, conventional dichotomous keys that need someone in the field team with the appropriate skills to use these highly technical keys. The lack of formal taxonomic teaching in the developing world and the demise of such teaching in the developed world means that increasingly people with the necessary skills are not available. This has led to significant problems in field collection or survey activities for conservation projects (notably with several CWR conservation projects funded by the Global Environment Facility), not to mention individual national activities around plant genetic resources. For example, the field botanists employed by the project “Conservation and Sustainable Use of Dryland Agrobiodiversity in Jordan, Lebanon, the Palestinian Authority and Syria” were contracted to undertake such field survey work but had little if any prior experience using traditional identification aids, which seriously limited project activities (Al-Atawneh et al. 2009). In this example, even when specific identification training was provided to the staff, few were sufficiently confident at the end of the course to go into the field and use conventional dichotomous keys to identify the project taxa. It forced the project team to rethink how field identification might best be achieved. The proposed solution was to create a series of user-friendly conservation field guides, the first of which was recently published by the International Center for Agricultural Research in the Dry Areas (ICARDA) for Medicago species (Al-Atawneh et al. 2009).
These conservation field guides contain a combination of simple traditional keys, descriptions, illustrations, photographs and interactive identification keys. A comparison of the basic difference between identification using a standard taxonomic Flora and conservation field guides is made in table 11.1. Using descriptions or illustrations for identification involves matching a particular specimen with the characteristics of known species that are drawn, photographed or described in the guide. Keys help users focus their search on a section of the guide where the number of choices is relatively small. Then, by scanning the species illustrations, a tentative identification can be made. The best guides are the ones that give references to similar species in each species account. Users can make direct comparisons with these to increase the confidence of a positive identification. Sometimes, one single taxon-specific character among all of those given is enough to identify the species (e.g., a leaf, a flower, a twig, a fruit, or a piece of bark for trees, or even a specific habitat).
Table 11.1. Comparison of floras and field guides
|
Flora |
Conservation Field Guide |
|
Designed to be used primarily in herbaria by taxonomists (cumbersome to take on field trips) |
Designed to be used primarily in the field by field botanists (robust structure suitable for field use) |
|
Rarely planned for use by non-experts |
Planned for use by non-experts, often commercial |
|
Heavy and expensive, occupying much shelf space |
Ideally in one portable volume |
|
Emphasizing formal taxonomy, with lists of synonyms and specimen citations |
Emphasizing information required to identify the plants |
|
Resolving issues over taxonomic limits, accepted plant names and synonyms |
Relying on other works (usually a Flora or monograph) to define accepted plant names |
|
Relying on traditional dichotomous keys and full descriptions for identification |
Using a wide range of identification aids |
|
Generally including all vascular plants in a geographic area |
Typically restricted to a narrower subset of all species than a flora (for example, crop wild relatives) |
|
Focus on precise botanical characters, even if obscure, but offering precision |
Focus on easily observed field characters |
|
Often take decades to write, lack practical information on conservation status, cultivation or usage |
Usually prepared relatively quickly and containing additional information like conservation status, cultivation or usage |
|
Few illustrations or pictures |
Most species illustrated or photographs included |
Source: Adapted from Lawrence and Hawthorn (2006).
What makes a “conservation” field guide? Many field guides have a focus on identification alone but a “conservation” field guide should enhance the conservation value of the guide by including information on current conservation status and threat assessment in the centre of diversity of the genus, along with future conservation requirements. Further, for conservationists of plant genetic resources, there is an essential link between conservation and use; therefore, for each species the guide would include additional information on the actual and potential use value of the species. There is no precise universal format for such a guide, especially as they may focus on a particular habitat, region or taxon, but they commonly present a taxonomic background, morphological descriptions, habitats, behaviour, ecology, distribution maps, uses, conservation notes and simple dichotomous keys suitable for field use, possibly annotated with line drawings, photographs, or paintings. To specifically address the problem of non-expert identification, the printed guide will ideally be accompanied by a CD with an interactive identification system to the taxa covered in the guide.
A good guide is one that is widely used by the target audience. It should be accurate, attractive, relevant, affordable and available; but it should also convey good-quality information that actually improves the knowledge of the user. Lawrence and Hawthorne (2006) define this as the guide being usable: users can find, understand and apply the information contained in it to meet their needs. Lawrence and Hawthorne stress that the authors of such guides need to involve a range of experts (who may include local experts with traditional knowledge), as well as potential users, in planning and researching the guide’s style and content so that the final product is the result of collaboration and not dictation.
A useful development would an on-line list of available conservation field guides. Further, such a site could be a location from which on-line conservation field guides could be downloaded, possibly similar to the eFloras website where copies of some standard Floras can be downloaded (www.efloras.org/index.aspx). This would mean that, as the resource grew, anyone planning conservation could look on-line at the site and download the identification tool they require for the taxa or region in which they are planning to undertake active conservation.
Barcode identification
In recent years, the rapid advance of molecular techniques has opened the possibility of using DNA samples for specimen identification (Kress et al. 2005), and so-called DNA barcoding uses a short genetic marker in an organism's DNA to identify it as belonging to a particular species. While this would not have a field application (at least at this time), it does mean that if a specimen cannot be identified using more conventional means, a DNA sample could be taken and compared against a DNA reference collection. Specimens can also be identified from plant parts (e.g., even leaves or roots) when the flowers or fruit normally used for identification are unavailable.
For the system to work, a desirable locus for DNA barcoding needs to be agreed upon, so that large databases of sequences for that locus can be developed for each plant species. The Plant Working Group for the Consortium for the Barcode of Life (CBOL) (2009) have suggested the concatenation of the rbcL and matK chloroplast genes as the locus for plants. The DNA sequences would be stored in a DNA sequence database, like GenBank, but there would be a need to link DNA sequences to vouchered specimens to ensure that the sequences are grounded to a named specimen of the taxon (Miller 2007). Although DNA barcoding has its critics as a technique and there are still technical problems that remain with its practical implementation, it seems likely to be a technique of growing importance in future years.
Accepted names
Identification is a two-stage process. First, the decision must be made as to which taxon (e.g., genus, species or variety) the specimen represents and, second, what the “accepted” name for that taxon is. Taxonomists’ views on the “accepted” name for a taxon will continue to vary as views on how taxa are related change with new taxonomic evidence. While this will occasionally result in taxon name changes, in recent years there has been significant progress toward the production of more stable lists of accepted species. The Convention on Biological Diversity (CBD) recognized the need for a stable list of taxa in the Global Strategy for Plant Conservation 2011-2020 (CBD 2010) under Objective I: “Plant diversity is well understood, documented and recognized”. The first specific action (Target 1) is an online Flora of all known plants.
Since the 1980s many multi-institutional database projects have been collating biodiversity information for either specific taxonomic groups or geographical regions. For example, the International Legume Database and Information Service (ILDIS) (http://ildis.org/) was one of the first monographic databases to be established for the 17,000 species of legumes (Zarucchi et al. 1993). This project is managed as a cooperative, involving approximately 20 research groups from five continents, and the information system is available internationally. Similar monographic projects have been established for several other plant groups (e.g., the Gymnosperm Database (www.conifers.org/) of taxonomic and specimen data for all 619 species of conifers in the world; the Brassicaceae species checklist and database, (www.cbif.gc.ca/pls/spec/brassicaceae) a species checklist of accepted names and synonyms for the Brassicaceae (Cruciferae) family; and Solanaceae Source (www.solanaceaesource.org), a database of names and descriptions of all taxa in the genus Solanum. There have also been floristic databases for regions, such as the Euro+Med PlantBase (www.emplantbase.org), which provides an on-line database and information system for the vascular plants of Europe and the Mediterranean region.
However, perhaps the most significant advance in recent years is The Plant List (www.theplantlist.org/), a collaboration between the Royal Botanic Gardens, Kew, in the UK; Missouri Botanical Gardens, St Louis, Missouri, in the USA; and other partners worldwide, to create the first working list of all known plants. It specifically addresses Target 1 of the Global Strategy for Plant Conservation for “a working list of known plant species” to be made available by 2010. It includes data on vascular plants (flowering plants, conifers, ferns and their allies) and of Bryophytes (mosses and liverworts). Currently it includes 1.25 million scientific plant names, of which 1.04 million are names of species rank. Of the species names included in The Plant List, about 300,000 (29%) are accepted names for species and about 480,000 (46%) are recorded as synonyms of those plant species. The status of the remaining 260,000 names is “unresolved” since the contributing data sets do not contain sufficient evidence to decide whether they should be accepted names or synonyms. The Plant List also includes a further 204,000 scientific plant names of infra-specific taxonomic rank linked to the species names.
Even more ambitious is the Catalogue of Life (www.catalogueoflife.org) project, developed by Species 2000 and the Integrated Taxonomic Information System (ITIS), which is planned to become a comprehensive catalogue of all known species of organisms on the planet. Rapid progress has been made recently in bringing together individual database projects, and the eleventh edition of the Annual Checklist has recently been published with 1,368,009 species. This is probably just slightly over two-thirds of the world's known species. The present Catalogue of Life is compiled from about 100 taxonomic databases from around the world. Many of these contain taxonomic data and opinions from extensive networks of specialists; the complete work contains contributions from more than 3000 specialists from throughout the taxonomic profession. The Species 2000 and ITIS teams peer review databases, select appropriate sectors and integrate the sectors into a single coherent catalogue with a single hierarchical classification. While the introduction of alternative taxonomic treatments and alternative classifications is planned, an important feature is that, for those users who wish to use it, a single preferred catalogue, based on peer reviews, will continue to be provided. It seems likely that in a few years we will take it for granted that such comprehensive lists of accepted names are available for our use, but the work involved in producing such lists should not be underestimated or go without being acknowledged.
Future challenges/needs/gaps
When the original text for this chapter was written, it would have been difficult to predict what challenges would be met and what gaps would remain, but clearly there have been advances—most notably in the use of molecular techniques to delimit and distinguish species and the production of a global list of accepted names. However, little if any progress has been made in moving beyond using traditional dichotomous keys as the routine basis for identification. Why? A cynic might point out that taxonomists are quite able to use traditional keys and have no real incentive to develop a more user-friendly means of identification, noting at the same time that both the use of molecular techniques and a global list of accepted names directly benefit their own taxonomic research. Yet there is no question that poor taxonomic training and lack of user-friendly identification aids are inhibiting the conservation and use of plant genetic resources. The gap remains, and the challenge will be to persuade funding agencies to address this gap. The most likely solution in the digital age is a mixture of text- and image-based interactive identification software that is taxa comprehensive and can run on cell phones or tablets that can easily be taken into the field. Allied to this, there remains a need for school and graduate training in applied taxonomy so that students and budding conservationists are not hobbled by deficiencies in their taxonomic skills.
Conclusion
Although there have been advances in identification science in recent years, poor taxonomic training and lack of user-friendly identification aids are currently inhibiting the conservation and use of plant genetic resources. This is resulting in (1) target species being missed in the field, (2) duplication of efforts in trying to locate priority taxa, (3) misidentification of conserved resources and (4) lack or poor utilization of conserved resources. Establishing time-bound targets and multi-institutional collaborative action has been shown to be very successful in generating the first working list of all known plant species. Could we learn from this example? Whatever approach is agreed upon, action is required now or our limited conservation resources will continue to be wasted. In the face of climate change and human mismanagement of the environment, can we continue to afford such waste?
Back to list of chapters on collecting
References and further reading
Al-Atawneh N, Shehadeh A, Amri A, Maxted N. 2009. Conservation Field Guide to Medics of the Mediterranean Basin. ICARDA, Aleppo, Syria.
Beentje H. 2010. The Kew Plant Glossary: An Illustrated Dictionary of Plant Terms. Royal Botanic Gardens, Kew, UK.
CBD. 2010. Global Strategy for Plant Conservation 2011-2020. CBD Secretariat, Ottawa, Canada (www.cbd.int/gspc/targets.shtml) .
CBOL Plant Working Group. 2009. A DNA barcode for land plants. Proceedings of the National Academy of Sciences 106(31):12794–12797.
FAO. 2010. Second Report on the State of the World’s Plant Genetic Resources for Food and Agriculture. FAO, Rome (www.fao.org/agriculture/seed/sow2/en/).
Feuillet C, Langridge P, Waugh R. 2008. Cereal breeding takes a walk on the wild side. Trends in Genetics 24:24–32.
Harris JG, Woolf Harris M. 2001. Plant Identification Terminology: An Illustrated Glossary. Spring Lake Publishing, Payson, Utah, USA.
Hickey M, King C. 2000. The Cambridge Illustrated Glossary of Botanical Terms. Cambridge University Press, Cambridge, UK.
House of Lords. 2002. What on Earth? The Threat to the Science Underpinning Conservation. Select Committee appointed to consider Science and Technology, House of Lords, London (www.publications.parliament.uk/pa/ld200102/ldselect/ldsctech/118/11802.htm).
Kress WJ, Wurdack KJ, Zimmer EA, Weigt LA, Janzen DH. 2005. Use of DNA barcodes to identify flowering plants. Proceedings of the National Academy of Sciences 102(23):8369–8374.
Lawrence A, Hawthorne W. 2006. Plant Identification: Creating User-Friendly Field Guides for Biodiversity Management. Earthscan, London.
Maxted N, Kell SP. 2009. Establishment of a Global Network for the In Situ Conservation of Crop Wild Relatives: Status and Needs. Background Study Paper No. 39. Commission on Genetic Resources for Food and Agriculture, FAO, Rome.
Maxted N, Mabuza-Dlamini P, Moss H, Padulosi S, Jarvis A, Guarino L. 2004. An ecogeographic survey: African Vigna. Systematic and Ecogeographic Studies of Crop Genepools 10. IPGRI, Rome.
Miller SE. 2007. DNA barcoding and the renaissance of taxonomy. Proceedings of the National Academy of Sciences 104(12):4775–4776.
Pankhurst RJ. 1991. Practical Taxonomic Computing. Cambridge University Press, Cambridge, UK.
Stevenson RD, Haber WA, Morris RA. 2003. Electronic field guides and user communities in the eco-informatics revolution. Conservation Ecology 7(1):3.
Zarucchi JL, Winfield PJ, Polhill RM, Hollis S, Bisby FA, Allkin R. 1993. The ILDIS Project on the world's legume species diversity. In: Bisby FA, Russell GF, Pankhurst RJ, editors. Designs for a Global Plant Species Information System. Oxford University Press, Oxford, UK. pp. 131–144.
Internet resources
Aids to Identification in Difficult Groups of Animals and Plants (AIDGAP): www.field-studies-council.org/publications/aidgaptesters.asp
Brassicaceae species checklist and database: www.cbif.gc.ca/pls/spec/brassicaceae
Catalogue of Life: www.catalogueoflife.org
eFloras: www.efloras.org/index.aspx
Euro+Med PlantBase: www.emplantbase.org
Gymnosperm Database: www.conifers.org/
International Legume Database and Information Service (ILDIS): http://ildis.org/
Linnaeus II: www.eti.uva.nl/products/linnaeus.php
Lucid: www.lucidcentral.com/
Solanaceae Source: www.solanaceaesource.org
The Plant List: www.theplantlist.org/
Comments
- No comments found




.jpg)

Leave your comments
Post comment as a guest